[NiFe] Group 1d-hydrogenase
This entry was last updated at: Feb. 9, 2018, 7:26 a.m.
Properties
| Group | [NiFe] Group 1: Respiratory H2-uptake [NiFe] hydrogenases |
| Subgroup | [NiFe] Group 1d: Oxygen-tolerant |
| Function | Hydrogenotrophic respiration using O2 or fumarate as terminal electron acceptors. Enzyme transfers H2-liberated electrons through cyt b and quinone to terminal reductase. |
| Activity | H2-uptake (unidirectional) |
| Oxygen tolerance | O2-tolerant |
| Localisation | Membrane-bound |
Distribution
| Ecosystem distribution | Diverse soil, aquatic, and host-associated environments |
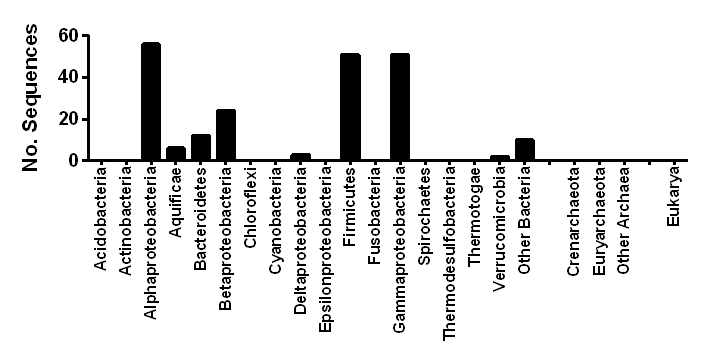
| Taxonomic distribution | Widespread among obligately aerobic and facultatively anaerobic bacteria |
Genetic Organisation
e.g. Escherichia coli (WP_021522685.1):

Architecture
| Structures |
|
| Subunits | 3 |
| Subunit description | HyaB (hydrogenase large subunit) HyaA (hydrogenase small subunit) HyaC (cytochrome b subunit) |
| Catalytic site | [NiFe]-centre |
| FeS clusters | Proximal: 6Cys[4Fe3S] Medial: 3Cys[3Fe4S] Distal: 3Cys1His[4Fe4S] |
Sequences in this class
| NCBI Accession | Organism | Hydrogenase Class | Phylum | Order | Activity (Predicted) | Oxygen Tolerance (Predicted) | Subunits (Predicted) | Metal Centres (Predicted) | Accessory Subunits (Predicted) |
|---|---|---|---|---|---|---|---|---|---|
| WP_008030831.1 | Rhodobacter sp. SW2 | [NiFe] Group 1d | Alphaproteobacteria | Rhodobacterales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_002720659.1 | Rhodobacter sphaeroides | [NiFe] Group 1d | Alphaproteobacteria | Rhodobacterales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_008282509.1 | Roseovarius sp. TM1035 | [NiFe] Group 1d | Alphaproteobacteria | Rhodobacterales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_005863657.1 | Sagittula stellata | [NiFe] Group 1d | Alphaproteobacteria | Rhodobacterales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_020041893.1 | Salipiger mucosus | [NiFe] Group 1d | Alphaproteobacteria | Rhodobacterales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_025825897.1 | Acetobacter okinawensis | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_025861579.1 | Acetobacter papayae | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_029009006.1 | Azospirillum halopraeferens | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_014249365.1 | Azospirillum lipoferum | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_009540200.1 | Caenispirillum salinarum | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_025813214.1 | Komagataeibacter kakiaceti | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_021130964.1 | Phaeospirillum fulvum | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_002725307.1 | Phaeospirillum molischianum | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_011388917.1 | Rhodospirillum rubrum | [NiFe] Group 1d | Alphaproteobacteria | Rhodospirillales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_020923409.1 | Rhizobium etli | [NiFe] Group 1d | Alphaproteobacteria | Rhozobiales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
| WP_008068262.1 | Novosphingobium nitrogenifigens | [NiFe] Group 1d | Alphaproteobacteria | Sphingomonadales | Aerobic Uptake | Tolerant | 3 | [NiFe]-centre, 1 x [4Fe3S] cluster, 1 x [3Fe4S] cluster, 1 x [4Fe4S] cluster | Cytochrome b |
Literature
Genetics:
- Bernhard, M., Schwartz, E., Rietdorf, J., and Friedrich, B. (1996) The Alcaligenes eutrophus membrane-bound hydrogenase gene locus encodes functions involved in maturation and electron transport coupling. J. Bacteriol. 178: 4522-4529.
- Brewin, N.J., DeJong, T.M., Phillips, D.A., and Johnston, A.W.B. (1980) Co-transfer of determinants for hydrogenase activity and nfodulation ability in Rhizobium leguminosarum. Nature 288: 77-79.
- Brito, B., Martínez, M., Fernández, D., Rey, L., Cabrera, E., Palacios, J.M., Imperial, J. and Ruiz-Argüeso, T. (1997) Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein NifA. Proc. Acad. Natl. Sci. USA 84: 6019-6024.
- Colbeau, A., Kóvacs, K.L., Chabert, J., and Vignais, P.M., 1994. Cloning and sequences of the structural (hupSLC) and accessory (hupDHI) genes for hydrogenase biosynthesis in Thiocapsa roseopersicina. Gene 140: 25-31.
- Csaki, R., Hanczar, T., Bodrossy, L., Murrell, J.C., and Kovács, K.L. (2001) Molecular characterization of structural genes coding for a membrane bound hydrogenase in Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 205: 203-207.
- Elsen, S., Dischert, W., Colbeau, A., and Bauer, C.E. (2000) Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J. Bacteriol. 182: 2831-2837.
- Ford, C.M., Garg, N., Garg, R.P., Tibelius, K.H., Yates, M.G., Arp, D.J., and Seefeldt, L.C. (1990) The identification, characterization, sequencing and mutagenesis of the genes (hupSL) encoding the small and large subunits of the H2-uptake hydrogenase of Azotobacter chroococcum. Mol. Microbiol. 4: 999-1008.
- Friedrich, B., Hogrefe, C., and Schlegel, H.G. (1981) Naturally occurring genetic transfer of hydrogen-oxidizing ability between strains of Alcaligenes eutrophus. J. Bacteriol. 147: 198-205.
- Greening, C., Biswas, A., Carere, C.R., Jackson, C.J., Taylor, M.C., Stott, M.B., Cook, G.M., and Morales, S.E. (2016) Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10: 761-777.
- Gutierrez, D., Hernando, Y., Palacios, J.M., Imperial, J., and Ruiz-Argüeso, T. (1997) FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum biovar viciae UPM791. J. Bacteriol. 179: 5264-5270.
- Jamieson, D.J., Sawers, R.G., Rugman, P.A., Boxer, D.H. and Higgins, C.F. (1986) Effects of anaerobic regulatory mutations and catabolite repression on regulation of hydrogen metabolism and hydrogenase isoenzyme composition in Salmonella typhimurium. J. Bacteriol. 168: 405-411.
- Leclerc, M., Colbeau, A., Cauvin, B., and Vignais, P.M. (1988) Cloning and sequencing of the genes encoding the large and the small subunits of the H2 uptake hydrogenase (hup) of Rhodobacter capsulatus. Mol. Gen. Genet. 214: 97-107.
- King, P.W. and Przybyla, A.E. (1999) Response of hya expression to external pH in Escherichia coli. J. Bacteriol. 181: 5250-5256.
- Lim, S.T. (1978) Determination of hydrogenase in free-living cultures of Rhizobium japonicum and energy efficiency of soybean nodules. Plant Physiol. 62: 609-611.
- Maier, R.J., Campbell, N.E., Hanus, F.J., Simpson, F.B., Russell, S.A., and Evans, H.J. (1978) Expression of hydrogenase activity in free-living Rhizobium japonicum. Proc. Acad. Natl. Sci. USA 75: 3258-3262.
- Menon, N.K., Robbins, J., Wendt, J.C., Shanmugam, K.T., and Przybyla, A.E. (1991) Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J. Bacteriol. 173: 4851-4861.
- Menon, N.K., Robbins, J., Peck, H.D., Chatelus, C.Y., Choi, E.S., and Przybyla, A.E. (1990) Cloning and sequencing of a putative Escherichia coli [NiFe] hydrogenase-1 operon containing six open reading frames. J. Bacteriol. 172: 1969-1977.
- Nelson, L.M. and Salminen, S.O. (1982) Uptake hydrogenase activity and ATP formation in Rhizobium leguminosarum bacteroids. J. Bacteriol. 151: 989-995.
- Palacios, J.M., Murillo, J., Leyva, A., Ditta, G. and Ruiz-Argüeso, T. (1990) Differential expression of hydrogen uptake (hup) genes in vegetative and symbiotic cells of Rhizobium leguminosarum. Mol. Gen. Genet. 221: 363-370.
- Pandelia, M.E., Lubitz, W., and Nitschke, W. (2012) Evolution and diversification of Group 1 [NiFe] hydrogenases. Is there a phylogenetic marker for O2-tolerance? BBA Bioenergetics 1817: 1565-1575.
- Sayavedra-Soto, L.A., Powell, G.K., Evans, H.J., and Morris, R.O. (1988) Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc. Natl. Acad. Sci. USA 85: 8395-8399.
- Schwartz, E., Henne, A., Cramm, R., Eitinger, T., Friedrich, B., and Gottschalk, G. (2003) Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based lithoautotrophy and anaerobiosis. J. Mol. Biol. 332: 369-383.
- Ueda, Y., Yamamoto, M., Urasaki, T., Arai, H., Ishii, M., and Igarashi, Y. (2007) Sequencing and reverse transcription-polymerase chain reaction (RT-PCR) analysis of four hydrogenase gene clusters from an obligately autotrophic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus TK-6. J. Biosci. Bioeng. 104: 470-475.
- Uffen, R.L., Colbeau, A., Richaud, P., and Vignais, P.M. (1990) Cloning and sequencing the genes encoding uptake-hydrogenase subunits of Rhodocyclus gelatinosus. Mol. Gen. Genet. 221: 49-58.
Physiology:
- Aguilar, O.M., Yates, M.G., and Postgate, J.R. (1985) The beneficial effect of hydrogenase in Azotobacter chroococcum under nitrogen-fixing, carbon-limiting conditions in continuous and batch cultures. Microbiol. 131: 3141-3145.
- Albrecht, S.L., Maier, R.J., Hanus, F.J., Russell, S.A., Emerich, D.W., and Evans, H.J. (1979) Hydrogenase in Rhizobium japonicum increases nitrogen fixation by nodulated soybeans. Science 203: 1255-1257.
- Ballantine, S.P. and Boxer, D.H. (1985) Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J. Bacteriol. 163: 454-459.
- Bowman, L., Balbach, J., Walton, J., Sargent, F., and Parkin, A. (2016) Biosynthesis of Salmonella enterica [NiFe]-hydrogenase-5: probing the roles of system-specific accessory proteins. J. Biol. Inorg. Chem. 21: 865-873.
- Brøndsted, L. and Atlung, T. (1994) Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J. Bacteriol. 176: 5423-5428.
- Brugna-Guiral, M, Tron, P, Nitschke, W., Stetter, K.-O., Burlat, B., Guigliarelli, B, Bruschi, M., and Giudici-Orticoni, M.T. (2003) [NiFe] hydrogenases from the hyperthermophilic bacterium Aquifex aeolicus: properties, function, and phylogenetics. Extremophiles 7: 147-157.
- Carere, C.R., Hards, K., Houghton, K.M., Power, J.F., McDonald, B., Collet, C., Gapes, D.J., Sparling, R., Boyd, E.S., Cook, G.M., Greening, C., and Stott, M.B. (2017) Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J. 11: 2599-2610.
- Colbeau, A., Kelley, B.C., and Vignais, P.M. (1980) Hydrogenase activity in Rhodopseudomonas capsulata: relationship with nitrogenase activity. J. Bacteriol. 144: 141-148.
- Colbeau, A. and Vignais, P.M. (1983) The membrane-bound hydrogenase of Rhodopseudomonas capsulata is inducible and contains nickel. BBA Prot. Struct. Mol. Enzymol. 748: 128-138.
- De Bont, J.A.M. (1976) Hydrogenase activity in nitrogen-fixing methane-oxidizing bacteria. Antonie van Leeuwenhoek 42: 255-259.
- Friedrich, C.G., Friedrich, B. and Bowien, B. (1981) Formation of enzymes of autotrophic metabolism during heterotrophic growth of Alcaligenes eutrophus. Microbiol. 122: 69-78.
- Guiral, M., Tron, P., Aubert, C., Gloter, A., Iobbi-Nivol, C., and Giudici-Orticoni, M.T. (2005) A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J. Biol. Chem. 280: 42004-42015.
- Guiral, M., Tron, P., Belle, V., Aubert, C., Léger, C., Guigliarelli, B., Giudici-Orticoni, M.T. (2006) Hyperthermostable and oxygen resistant hydrogenases from a hyperthermophilic bacterium Aquifex aeolicus: physicochemical properties. Int. J. Hydrogen Energy 31: 1424-1431.
- Hanczár, T., Csáki, R., Bodrossy, L., Murrell, C.J., and Kovács, K.L. (2002) Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch. Microbiol. 177: 167-172.
- Hanus, F.J., Maier, R.J., and Evans, H.J. (1979) Autotrophic growth of H2-uptake-positive strains of Rhizobium japonicum in an atmosphere supplied with hydrogen gas. Proc. Acad. Natl. Sci. USA 76: 1788-1792.
- Hedrich, S. and Johnson, D.B. (2013) Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol. Lett. 349: 40-45.
- Kávacs, K.L., Kávacs, A.T., Maroti, G., Meszaros, L.S., Balogh, J., Latinovics, D., Fülöp, A., David, R., Doroghazi, E., and Rákhely, G. (2005) The hydrogenases of Thiocapsa roseopersicina. Biochem. Soc. Trans. 33: 61-63.
- Lamichhane-Khadka, R., Benoit, S.L., Miller-Parks, E.F., and Maier, R.J., 2015. Host hydrogen rather than that produced by the pathogen is important for Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 83: 311-316.
- Laurinavichene, T.V. and Tsygankov, A.A. (2001) H2 consumption by Escherichia coli coupled via hydrogenase 1 or hydrogenase 2 to different terminal electron acceptors. FEMS Microbiol. Lett. 202: 121-124.
- Laurinavichene, T.V., Zorin, N.A., and Tsygankov, A.A. (2002) Effect of redox potential on activity of hydrogenase 1 and hydrogenase 2 in Escherichia coli. Arch. Microbiol. 178: 437-442.
- McNorton, M.M. and Maier, R.J. (2012) Roles of H2 uptake hydrogenases in Shigella flexneri acid tolerance. Microbiol. 158: 2204-2212.
- Mohammadi, S., Pol, A., van Alen, T.A., Jetten, M.S., and den Camp, H.J.O. (2017) Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and-insensitive hydrogenase. ISME J. 11: 945-958.
- Pálágyi-Mészáros, L.S., Maróti, J., Latinovics, D., Balogh, T., Klement, E., Medzihradszky, K.F., Rákhely, G., and Kovács, K.L. (2009) Electron-transfer subunits of the NiFe hydrogenases in Thiocapsa roseopersicina BBS. FEBS J. 276: 164-174.
- Pedrosa, F.O., Stephan, M., Dobereiner, J., and Yates, M.G. (1982) Hydrogen-uptake hydrogenase activity in nitrogen-fixing Azospirillum brasilense. Microbiol. 128: 161-166.
- Pinske, C., McDowall, J.S., Sargent, F., and Sawers, R.G. (2012) Analysis of hydrogenase 1 levels reveals an intimate link between carbon and hydrogen metabolism in Escherichia coli K-12. Microbiol. 158: 856-868.
- Richard, D.J., Sawers, G., Sargent, F., McWalter, L., and Boxer, D.H. (1999) Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiol. 145: 2903-2912.
- Sato, T., Inoue, K., Sakurai, H., and Nagashima, K.V. (2017) Effects of the deletion of hup genes encoding the uptake hydrogenase on the activity of hydrogen production in the purple photosynthetic bacterium Rubrivivax gelatinosus IL144. J. Gen. Appl. Microbiol. 63: 274-279.
- Sawers, R.G., Jamieson, D.J., Higgins, C.F., and Boxer, D.H. (1986) Characterization and physiological roles of membrane-bound hydrogenase isoenzymes from Salmonella typhimurium. J. Bacteriol. 168: 398-404.
- Shah, N.N., Hanna, M.L., Jackson, K.J., and Taylor, R.T. (1995) Batch cultivation of Methylosinus trichosporium OB3B: IV. Production of hydrogen-driven soluble or particulate methane monooxygenase activity. Biotechnol. Bioengineer. 45: 229-238.
- Santiago, B. and Meyer, O. (1997) Purification and molecular characterization of the H2 uptake membrane-bound NiFe-hydrogenase from the carboxidotrophic bacterium Oligotropha carboxidovorans. J. Bacteriol. 179: 6053-6060.
- Schink, B. (1982) Isolation of a hydrogenase-cytochrome b complex from cytoplasmic membranes of Xanthobacter autotrophicus GZ 29. FEMS Microbiol. Lett. 13: 289-293.
- Sim, E. and Vignais, P.M. (1978) Hydrogenase activity in Paracoccus denitrificans. Partial purification and interaction with the electron transport chain. Biochimie 60: 307-314.
- Uffen, R.L. and Wolfe, R.S. (1970) Anaerobic growth of purple nonsulfur bacteria under dark conditions. J. Bacteriol. 104: 462-472.
- Wong, T.Y. and Maier, R.J. (1985) H2-dependent mixotrophic growth of N2-fixing Azotobacter vinelandii. J. Bacteriol. 163: 528-533.
- Yates, M.G. and Robson, R.L. (1985) Mutants of Azotobacter chroococcum defective in hydrogenase activity. Microbiol. 131: 1459-1466.
- Zbell, A.L., Benoit, S.L., and Maier, R.J. (2007) Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiol. 153: 3508-3516.
- Zbell, A.L., Maier, S.E., and Maier, R.J. (2008) Salmonella enterica serovar Typhimurium NiFe uptake-type hydrogenases are differentially expressed in vivo. Infect. Immun. 76: 4445-4454.
- Zbell, A.L. and Maier, R.J. (2009) Role of the Hya hydrogenase in recycling of anaerobically produced H2 in Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 75: 1456-1459.
Biochemistry:
- Adams, M.W. and Hall, D.O. (1979) Purification of the membrane-bound hydrogenase of Escherichia coli. Biochem. J. 183: 11-22.
- Adams, M.W.W. and Hall, D.O. (1977) Isolation of the membrane-bound hydrogenase from Rhodospirillum rubrum. Biochem. Biophys. Res. Commun. 77: 730-737.
- Adams, M.W. and Hall, D.O. (1979) Properties of the solubilized membrane-bound hydrogenase from the photosynthetic bacterium Rhodospirillum rubrum. Arch. Biochem. Biophys. 195: 288-299.
- Adamson, H., Robinson, M., Wright, J.J., Flanagan, L.A., Walton, J., Elton, D., Gavaghan, D.J., Bond, A.M., Roessler, M.M., and Parkin, A. (2017) Retuning the catalytic bias and overpotential of a [NiFe]-hydrogenase via a single amino acid exchange at the electron entry/exit site. J. Am. Chem. Soc. 139: 10677-10686.
- Arp, D.J., McCollum, L.C., and Seefeldt, L.C. (1985) Molecular and immunological comparison of membrane-bound, H2-oxidizing hydrogenases of Bradyrhizobium japonicum, Alcaligenes eutrophus, Alcaligenes latus, and Azotobacter vinelandii. J. Bacteriol. 163: 15-20.
- Bernhard, M., Benelli, B., Hochkoeppler, A., Zannoni, D., and Friedrich, B. (1997) Functional and structural role of the cytochrome b subunit of the membrane-bound hydrogenase complex of Alcaligenes eutrophus H16. Eur. J. Biochem. 248: 179-186.
- Bowman, L., Flanagan, L., Fyfe, P.K., Parkin, A., Hunter, W.N., and Sargent, F. (2014) How the structure of the large subunit controls function in an oxygen-tolerant [NiFe]-hydrogenase. Biochem. J. 458: 449-458.
- Bürstel, I., Siebert, E., Frielingsdorf, S., Zebger, I., Friedrich, B., and Lenz, O. (2016) CO synthesized from the central one-carbon pool as source for the iron carbonyl in O2-tolerant [NiFe]-hydrogenase. Proc. Natl. Acad. Sci. USA 113: 14722-14726.
- Brooke, E.J., Evans, R.M., Islam, S.T., Roberts, G.M., Wehlin, S.A., Carr, S.B., Phillips, S.E., and Armstrong, F.A. (2016) Importance of the active site “canopy” residues in an O2-tolerant [NiFe]-hydrogenase. Biochem. 56: 132-142.
- Colbeau, A., Chabert, J. and Vignais, P.M. (1983) Purification, molecular properties and localization in the membrane of the hydrogenase of Rhodopseudomonas capsulata. BBA Prot. Struct. Mol. Enzymol. 748: 116-127.
- Cracknell, J.A., Wait, A.F., Lenz, O., Friedrich, B., and Armstrong, F.A. (2009) A kinetic and thermodynamic understanding of O2 tolerance in [NiFe]-hydrogenases. Proc. Natl. Acad. Sci. USA 106: 20681-20686.
- Eguchi, S., Yoon, K.S. and Ogo, S. (2012) O2-stable membrane-bound [NiFe] hydrogenase from a newly isolated Citrobacter sp. S-77. J. Biosci. Bioengin. 114: 479-484.
- Evans, R.M., Parkin, A., Roessler, M.M., Murphy, B.J., Adamson, H., Lukey, M.J., Sargent, F., Volbeda, A., Fontecilla-Camps, J.C., and Armstrong, F.A. (2013) Principles of sustained enzymatic hydrogen oxidation in the presence of oxygen-the crucial influence of high potential Fe-S clusters in the electron relay of [NiFe]-hydrogenases. J. Am. Chem. Soc. 135: 2694-2707.
- Flanagan, L.A., Wright, J.J., Roessler, M.M., Moir, J.W., and Parkin, A. (2016) Re-engineering a NiFe hydrogenase to increase the H2 production bias while maintaining native levels of O2 tolerance. Chem. Commun. 52: 9133-9136.
- Frielingsdorf, S. et al. (2014). Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase. Nature Chem. Biol. 10: 378-385.
- Frielingsdorf, S., Schubert, T., Pohlmann, A., Lenz, O., and Friedrich, B. (2011) A trimeric supercomplex of the oxygen-tolerant membrane-bound [NiFe]-hydrogenase from Ralstonia eutropha H16. Biochem. 50: 10836-10843.
- Fritsch, J., Lenz, O., and Friedrich, B. (2013) Structure, function and biosynthesis of O2-tolerant hydrogenases. Nature Rev. Microbiol. 11: 106-114.
- Fritsch, J., Scheerer, P., Frielingsdorf, S., Kroschinsky, S., Friedrich, B., Lenz, O., and Spahn, C.M. (2011) The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature 479: 249-252.
- Fritsch, J., Lenz, O., and Friedrich, B. (2011) The maturation factors HoxR and HoxT contribute to oxygen tolerance of membrane-bound [NiFe] hydrogenase in Ralstonia eutropha H16. J. Bacteriol. 193: 2487-2497.
- Goldet, G., Wait, A.F., Cracknell, J.A., Vincent, K.A., Ludwig, M., Lenz, O., Friedrich, B., and Armstrong, F.A. (2008) Hydrogen production under aerobic conditions by membrane-bound hydrogenases from Ralstonia species. J. Am. Chem. Soc. 130: 11106-11113.
- Goris, T., Wait, A.F., Saggu, M., Fritsch, J., Heidary, N., Stein, M., Zebger, I., Lendzian, F., Armstrong, F.A., Friedrich, B., and Lenz, O. (2011) A unique iron-sulfur cluster is crucial for oxygen tolerance of a [NiFe]-hydrogenase. Nature Chem. Biol. 7: 310-318.
- Guiral, M., Tron, P., Belle, V., Aubert, C., Léger, C., Guigliarelli, B., and Giudici-Orticoni, M.T. (2006) Hyperthermostable and oxygen resistant hydrogenases from a hyperthermophilic bacterium Aquifex aeolicus: physicochemical properties. Int. J. Hydrogen Energy 31: 1424-1431.
- Kalms, J., Schmidt, A., Frielingsdorf, S., van der Linden, P., von Stetten, D., Lenz, O., Carpentier, P., and Scheerer, P. (2016) Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase reveals a hydrophobic tunnel network for gas transport. Angewandte Chemie 55: 5586-5590.
- Knüttel, K. Schneider, K., Schlegel, H.G., and Müller, A. (1989) The membrane-bound hydrogenase from Paracoccus denitrificans. Eur. J. Biochem. 179: 101-108.
- Ludwig, M., Cracknell, J.A., Vincent, K.A., Armstrong, F.A., and Lenz, O. (2009) Oxygen-tolerant H2 oxidation by membrane-bound [NiFe] hydrogenases of Ralstonia species coping with low level H2 in air. J. Biol. Chem. 284: 465-477.
- Lukey, M.J., Parkin, A., Roessler, M.M., Murphy, B.J., Harmer, J., Palmer, T., Sargent, F., and Armstrong, F.A. (2010). How Escherichia coli is equipped to oxidize hydrogen under different redox conditions. J. Biol. Chem. 285: 3928-3938.
- Lukey, M.J., Roessler, M.M., Parkin, A., Evans, R.M., Davies, R.A., Lenz, O., Friedrich, B., Sargent, F., and Armstrong, F.A. (2011) Oxygen-tolerant [NiFe]-hydrogenases: the individual and collective importance of supernumerary cysteines at the proximal Fe-S cluster. J. Am. Chem. Soc. 133: 16881-16892.
- Madigan, M.T. and Gest, H. (1979) Growth of the photosynthetic bacterium Rhodopseudomonas capsulata chemoautotrophically in darkness with H2 as the energy source. J. Bacteriol. 137: 524-530.
- Murphy, B.J., Sargent, F., and Armstrong, F.A. (2014) Transforming an oxygen-tolerant [NiFe] uptake hydrogenase into a proficient, reversible hydrogen producer. Energy Environ. Sci. 7: 1426-1433.
- Pandelia, M.E., Fourmond, V., Tron-Infossi, P., Lojou, E., Bertrand, P., Léger, C., Giudici-Orticoni, M.T., and Lubitz, W. (2010) Membrane-bound hydrogenase I from the hyperthermophilic bacterium Aquifex aeolicus: enzyme activation, redox intermediates and oxygen tolerance. J. Am. Chem. Soc. 132: 6991-7004.
- Parkin, A., Bowman, L., Roessler, M.M., Davies, R.A., Palmer, T., Armstrong, F.A., and Sargent, F. (2012) How Salmonella oxidises H2 under aerobic conditions. FEBS Lett. 586: 536-544.
- Radu, V., Frielingsdorf, S., Evans, S.D., Lenz, O., and Jeuken, L.J. (2014) Enhanced oxygen-tolerance of the full heterotrimeric membrane-bound [NiFe]-hydrogenase of Ralstonia eutropha. J. Am. Chem. Soc. 136: 8512-8515.
- Schink, B. and Schlegel, H.G. (1979) The membrane-bound hydrogenase of Alcaligenes eutrophus. I. Solubilization, purification, and biochemical properties. BBA Enzymol. 567: 315-324.
- Seefeldt, L.C. and Arp, D.J. (1986) Purification to homogeneity of Azotobacter vinelandii hydrogenase: a nickel and iron containing αβ dimer. Biochimie 68: 25-34.
- Sezer, M., Frielingsdorf, S., Millo, D., Heidary, N., Utesch, T., Mroginski, M.A., Friedrich, B., Hildebrandt, P., Zebger, I., and Weidinger, I.M. (2011) Role of the HoxZ subunit in the electron transfer pathway of the membrane-bound [NiFe]-hydrogenase from Ralstonia eutropha immobilized on electrodes. J. Phys. Chem. B 115: 10368-10374.
- Shomura, Y., Yoon, K.S., Nishihara, H., and Higuchi, Y. (2011) Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature 479: 253-256.
- So, K., Hamamoto, R., Takeuchi, R., Kitazumi, Y., Shirai, O., Endo, R., Nishihara, H., Higuchi, Y., and Kano, K., 2016. Bioelectrochemical analysis of thermodynamics of the catalytic cycle and kinetics of the oxidative inactivation of oxygen-tolerant [NiFe]-hydrogenase. J. Electroanalytical Chem. 766: 152-161.
- Vincent, K.A., Cracknell, J.A., Lenz, O., Zebger, I., Friedrich, B., and Armstrong, F.A. (2005) Electrocatalytic hydrogen oxidation by an enzyme at high carbon monoxide or oxygen levels. Proc. Acad. Natl. Sci. USA 102: 16951-16954.
- Vincent, K.A., Parkin, A., Lenz, O., Albracht, S.P., Fontecilla-Camps, J.C., Cammack, R., Friedrich, B., and Armstrong, F.A. (2005) Electrochemical definitions of O2 sensitivity and oxidative inactivation in hydrogenases. J. Am. Chem. Soc. 127: 18179-18189.
- Volbeda, A., Amara, P., Darnault, C., Mouesca, J.M., Parkin, A., Roessler, M.M., Armstrong, F.A., and Fontecilla-Camps, J.C. (2012) X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli. Proc. Natl. Acad. Sci. USA 109: 5305-5310.
- Volbeda, A., Darnault, C., Parkin, A., Sargent, F., Armstrong, F.A., and Fontecilla-Camps, J.C. (2013) Crystal structure of the O2-tolerant membrane-bound hydrogenase 1 from Escherichia coli in complex with its cognate cytochrome b. Structure 21: 184-190.
- Wulff, P., Day, C.C., Sargent, F., and Armstrong, F.A. (2014) How oxygen reacts with oxygen-tolerant respiratory [NiFe]-hydrogenases. Proc. Natl. Acad. Sci. USA 111: 6606-6611.
- Wulff, P., Thomas, C., Sargent, F., and Armstrong, F.A. (2016) How the oxygen tolerance of a [NiFe]-hydrogenase depends on quaternary structure. J. Biol. Inorg. Chem. 21: 121-134.
- Yoon, K.S., Fukuda, K., Fujisawa, K., and Nishihara, H. (2011) Purification and characterization of a highly thermostable, oxygen-resistant, respiratory [NiFe]-hydrogenase from a marine, aerobic hydrogen-oxidizing bacterium Hydrogenovibrio marinus. Int. J. Hydrogen Energy 36: 7081-7088.
Chemistry:
- Fritsch, J. et al. (2011) [NiFe] and [FeS] cofactors in the membrane-bound hydrogenase of Ralstonia eutropha investigated by X-ray absorption spectroscopy: insights into O2-tolerant H2 cleavage. Biochem. 50: 5858-5869.
- Pandelia, M.E., Bykov, D., Izsak, R., Infossi, P., Giudici-Orticoni, M.T., Bill, E., Neese, F., and Lubitz, W. (2013) Electronic structure of the unique [4Fe-3S] cluster in O2-tolerant hydrogenases characterized by 57Fe Mössbauer and EPR spectroscopy. Proc. Natl. Acad. Sci. USA 110: 483-488.
- Pandelia, M.E., Infossi, P., Giudici-Orticoni, M.T., and Lubitz, W. (2010) The oxygen-tolerant hydrogenase I from Aquifex aeolicus weakly interacts with carbon monoxide: an electrochemical and time-resolved FTIR study. Biochem. 49: 8873-8881.
- Pandelia, M.E., Infossi, P., Stein, M., Giudici-Orticoni, M.T., and Lubitz, W. (2012) Spectroscopic characterization of the key catalytic intermediate Ni-C in the O2-tolerant [NiFe] hydrogenase I from Aquifex aeolicus: evidence of a weakly bound hydride. Chem. Commun. 48: 823-825.
- Radu, V., Frielingsdorf, S., Lenz, O., and Jeuken, L.J. (2016) Reactivation from the Ni-B state in [NiFe] hydrogenase of Ralstonia eutropha is controlled by reduction of the superoxidised proximal cluster. Chem. Commun. 52: 2632-2635.
- Roessler, M.M., Evans, R.M., Davies, R.A., Harmer, J., and Armstrong, F.A. (2012) EPR spectroscopic studies of the Fe-S clusters in the O2-tolerant [NiFe]-hydrogenase Hyd-1 from Escherichia coli and characterization of the unique [4Fe-3S] cluster by HYSCORE. J. Am. Chem. Soc. 134: 15581-15594.
- Saggu, M., Teutloff, C., Ludwig, M., Brecht, M., Pandelia, M.E., Lenz, O., Friedrich, B., Lubitz, W., Hildebrandt, P., Lendzian, F., and Bittl, R. (2010) Comparison of the membrane-bound [NiFe] hydrogenases from R. eutropha H16 and D. vulgaris Miyazaki F in the oxidized ready state by pulsed EPR. Phys. Chem. Chem. Phys. 12: 2139-2148.
- Saggu, M., Zebger, I., Ludwig, M., Lenz, O., Friedrich, B., Hildebrandt, P., and Lendzian, F. (2009) Spectroscopic insights into the oxygen-tolerant membrane-associated [NiFe] hydrogenase of Ralstonia eutropha H16. J. Biol. Chem. 284: 16264-16276.
- Siebert, E. et al. (2015) Resonance Raman spectroscopic analysis of the [NiFe] active site and the proximal [4Fe-3S] cluster of an O2-tolerant membrane-bound hydrogenase in the crystalline state. J. Phys. Chem. 119: 13785-13796.
- Wisitruangsakul, N., Lenz, O., Ludwig, M., Friedrich, B., Lendzian, F., Hildebrandt, P., and Zebger, I. (2009) Monitoring catalysis of the membrane-bound hydrogenase from Ralstonia eutropha H16 by surface-enhanced IR absorption spectroscopy. Angewandte Chemie 48: 611-613.