[NiFe] Group 4f-hydrogenase Putative
This entry was last updated at: June 13, 2016, 11:25 a.m.
Properties
| Group | [NiFe] Group 4: Respiratory H2-evolving [NiFe] hydrogenases |
| Subgroup | [NiFe] Group 4f: Formate-coupled |
| Function | Unconfirmed role. May form respiratory complex that couples oxidation of formate to proton reduction. The energy generated is likely to be used to translocate protons through the antiporter-like subunits. |
| Activity | H2-evolution? |
| Oxygen tolerance | O2-sensitive |
| Localisation | Membrane-bound |
Distribution
| Ecosystem distribution | In low abundance in anoxic soils |
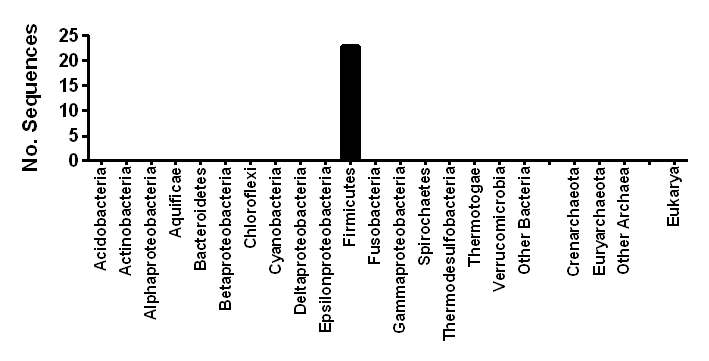
| Taxonomic distribution | Encoded in several classes of Firmicutes |
Genetic Organisation
e.g. Desulfosporosinus orientis (WP_014183752.1):

Architecture
| Structures |
|
| Subunits | 6? |
| Subunit description | EhfE (hydrogenase large subunit) EhfF (hydrogenase small subunit) EhfB (NuoH-like transmembrane adaptor) EhfA (NuoL-like transmembrane adaptor) EhfCD (antiporter-like subunits) |
| Catalytic site | [NiFe]-centre |
| FeS clusters | Proximal: 4Cys[4Fe4S] Medial: Absent Distal: Absent |
Important Notes
This hydrogenase class has only been identified based on genomic predictions. Physiological, genetic, and biochemical studies are needed to confirm its electron donor and describe its properties.
Sequences in this class
| NCBI Accession | Organism | Hydrogenase Class | Phylum | Order | Activity (Predicted) | Oxygen Tolerance (Predicted) | Subunits (Predicted) | Metal Centres (Predicted) | Accessory Subunits (Predicted) |
|---|---|---|---|---|---|---|---|---|---|
| WP_018133527.1 | Alicyclobacillus pohliae | [NiFe] Group 4f | Firmicutes | Bacillales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_006323220.1 | Anoxybacillus flavithermus | [NiFe] Group 4f | Firmicutes | Bacillales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_028986298.1 | Thermicanus aegyptius | [NiFe] Group 4f | Firmicutes | Bacillales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_005582638.1 | Clostridium ultunense | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_014795396.1 | Desulfitobacterium dehalogenans | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_015945149.1 | Desulfitobacterium hafniense | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_019851221.1 | Desulfitobacterium sp. PCE1 | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_014902103.1 | Desulfosporosinus meridiei | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_014183752.1 | Desulfosporosinus orientis | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_009619272.1 | Desulfosporosinus sp. OT | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_007780611.1 | Desulfosporosinus youngiae | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_015756422.1 | Desulfotomaculum acetoxidans | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_013843216.1 | Desulfotomaculum ruminis | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_013623369.1 | Syntrophobotulus glycolicus | [NiFe] Group 4f | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_004094701.1 | Acetonema longum | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_027938372.1 | Anaeroarcus burkinensis | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_018702624.1 | Anaeromusa acidaminophila | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_027397032.1 | Anaerovibrio lipolyticus | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_027406274.1 | Anaerovibrio sp. RM50 | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_021718870.1 | Phascolarctobacterium sp. CAG:207 | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_021167282.1 | Sporomusa ovata | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_019880045.1 | Succinispira mobilis | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
| WP_019553945.1 | Zymophilus raffinosivorans | [NiFe] Group 4f | Firmicutes | Selenomadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, two antiporter subunits |
Literature
Genetics:
- Greening, C., Biswas, A., Carere, C.R., Jackson, C.J., Taylor, M.C., Stott, M.B., Cook, G.M., and Morales, S.E. (2016) Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10: 761-777.