[NiFe] Group 4e-hydrogenase
This entry was last updated at: June 13, 2016, 11:11 a.m.
Properties
| Group | [NiFe] Group 4: Respiratory H2-evolving [NiFe] hydrogenases |
| Subgroup | [NiFe] Group 4e: Ferredoxin-coupled, Ech-type |
| Function | Forms respiratory complex that couples oxidation of ferredoxinred to reduction of protons to H2 in acetoclastic methanogenesis. This process appears to be coupled to sodium or proton translocation through the transmembrane modules. The complex can also act in the reverse direction during hydrogenotrophic methanogenesis. |
| Activity | Bidirectional |
| Oxygen tolerance | O2-sensitive |
| Localisation | Membrane-bound |
Distribution
| Ecosystem distribution | Anoxic soils and sediments, animal gastrointestinal tracts |
| Taxonomic distribution | Encoded in multiple methanogenic orders and anaerobic bacteria |
Genetic Organisation
e.g. Methanosarcina barkeri (WP_011305188.1):

Architecture
| Structures |
|
| Subunits | 6 |
| Subunit description | EchA (hydrogenase large subunit) EchC (hydrogenase small subunit) EchF (iron-sulfur subunit) EchB (NuoH-like transmembrane adaptor) EchA (NuoL-like transmembrane adaptor) EchD (NuoC-like transmembrane adaptor) |
| Catalytic site | [NiFe]-centre |
| FeS clusters | Proximal: 4Cys[4Fe4S] Medial: Absent Distal: Absent |
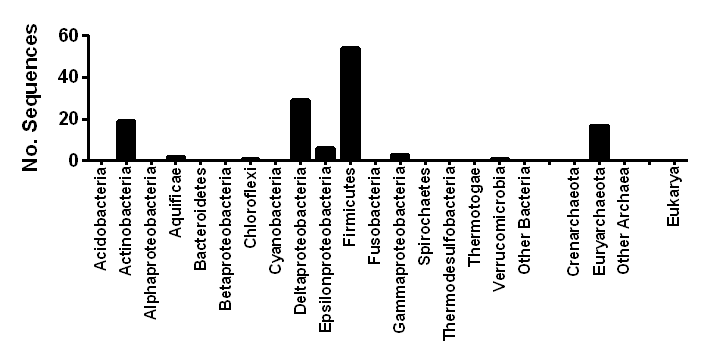
Sequences in this class
| NCBI Accession | Organism | Hydrogenase Class | Phylum | Order | Activity (Predicted) | Oxygen Tolerance (Predicted) | Subunits (Predicted) | Metal Centres (Predicted) | Accessory Subunits (Predicted) |
|---|---|---|---|---|---|---|---|---|---|
| WP_011937735.1 | Geobacter uraniireducens | [NiFe] Group 4e | Deltaproteobacteria | Desulfuromonadales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_013460916.1 | Sulfuricurvum kujiense | [NiFe] Group 4e | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_015654365.1 | Sulfuricurvum sp. RIFRC-1 | [NiFe] Group 4e | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_008340210.1 | Sulfurimonas gotlandica | [NiFe] Group 4e | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_007474009.1 | Caminibacter mediatlanticus | [NiFe] Group 4e | Epsilonproteobacteria | Nautiliales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_024789181.1 | Lebetimonas sp. JH292 | [NiFe] Group 4e | Epsilonproteobacteria | Nautiliales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012663581.1 | Nautilia profundicola | [NiFe] Group 4e | Epsilonproteobacteria | Nautiliales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012035507.1 | Methanocella arvoryzae | [NiFe] Group 4e | Euryarchaeota | Methanocellales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_014405402.1 | Methanocella conradii | [NiFe] Group 4e | Euryarchaeota | Methanocellales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012899275.1 | Methanocella paludicola | [NiFe] Group 4e | Euryarchaeota | Methanocellales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012899230.1 | Methanocella paludicola | [NiFe] Group 4e | Euryarchaeota | Methanocellales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_014867437.1 | Methanoculleus bourgensis | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_011843203.1 | Methanoculleus marisnigri | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_004039320.1 | Methanofollis liminatans | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_020449518.1 | Methanomassiliicoccus intestinalis | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_019176382.1 | Methanomassiliicoccus luminyensis | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_004077817.1 | Methanoplanus limicola | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_013328417.1 | Methanoplanus petrolearius | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_011991040.1 | Methanoregula boonei | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_015284201.1 | Methanoregula formicica | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012619310.1 | Methanosphaerula palustris | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_011448732.1 | Methanospirillum hungatei | [NiFe] Group 4e | Euryarchaeota | Methanomicrobiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_011305188.1 | Methanosarcina barkeri | [NiFe] Group 4e | Euryarchaeota | Methanosarcinales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_011034248.1 | Methanosarcina mazei | [NiFe] Group 4e | Euryarchaeota | Methanosarcinales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_010251151.1 | Acetivibrio cellulolyticus | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_015738579.1 | Ammonifex degensii | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_027399701.1 | Anaerovorax odorimutans | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_027203889.1 | Butyrivibrio fibrisolvens | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_026651247.1 | Butyrivibrio proteoclasticus | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_026892028.1 | Clostridium aerotolerans | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_005213155.1 | Clostridium celatum | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_027631079.1 | Clostridium cellobioparum | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_015926708.1 | Clostridium cellulolyticum | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_014254276.1 | Clostridium clariflavum | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_024832561.1 | Clostridium josui | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_024346172.1 | Clostridium methoxybenzovorans | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_004618281.1 | Clostridium papyrosolvens | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_004628623.1 | Clostridium termitidis | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_019226287.1 | Dehalobacter sp. E1 | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_020490799.1 | Dehalobacter sp. FTH1 | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012302199.1 | Desulforudis audaxviator | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_027871860.1 | Eubacterium cellulosolvens | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_026835345.1 | Eubacterium xylanophilum | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_012199760.1 | Lachnoclostridium phytofermentans | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_027431522.1 | Lachnospira multipara | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_028241974.1 | Pseudobutyrivibrio ruminis | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_028246016.1 | Pseudobutyrivibrio ruminis | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_028234750.1 | Pseudobutyrivibrio sp. MD2005 | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_003514863.1 | Ruminiclostridium thermocellum | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
| WP_007047619.1 | Subdoligranulum variabile | [NiFe] Group 4e | Firmicutes | Clostridiales | Evolving | Sensitive | 6 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein |
Literature
Genetics:
- Biswas, R., Zheng, T., Olson, D.G., Lynd, L.R., and Guss, A.M. (2015) Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol. Biofuels 8: 20.
- Calusinska, M., Happe, T., Joris, B., and Wilmotte, A. (2010) The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiol. 156: 1575-1588.
- Friedrich, T. and Scheide, D. (2000) The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479: 1-5.
- Greening, C., Biswas, A., Carere, C.R., Jackson, C.J., Taylor, M.C., Stott, M.B., Cook, G.M., and Morales, S.E. (2016) Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10: 761-777.
- Hedderich, R. (2004) Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembranes 36: 65-75.
- Marreiros, B.C., Batista, A.P., Duarte, A.M., and Pereira, M.M. (2013) A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. BBA Bioenerg. 1827: 198-209.
- Plugge, C.M., Scholten, J.C., Culley, D.E., Nie, L., Brockman, F.J., and Zhang, W. (2010) Global transcriptomics analysis of the Desulfovibrio vulgaris change from syntrophic growth with Methanosarcina barkeri to sulfidogenic metabolism. Microbiol. 156: 2746-2756.
- Rodrigues, R., Valente, F.M., Pereira, I.A., Oliveira, S., and Rodrigues-Pousada, C. (2003) A novel membrane-bound Ech [NiFe] hydrogenase in Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 306: 366-375.
Physiology:
- Guss, A.M., Mukhopadhyay, B., Zhang, J.K., and Metcalf, W.W. (2005) Genetic analysis of mch mutants in two Methanosarcina species demonstrates multiple roles for the methanopterin-dependent C-1 oxidation/reduction pathway and differences in H2 metabolism between closely related species. Mol. Microbiol. 55: 1671-1680.
- Hedderich, R. and Forzi, L. (2006) Energy-converting [NiFe] hydrogenases: more than just H2 activation. J. Mol. Microbiol. Biotechnol. 10: 92-104.
- Künkel, A., Vorholt, J.A., Thauer, R.K., and Hedderich, R. (1998) An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur. J. Biochem. 252: 467-476.
- Meuer, J., Kuettner, H.C., Zhang, J.K., Hedderich, R., and Metcalf, W.W. (2002) Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. USA 99: 5632-5637.
- Morais-Silva, F.O., Santos, C.I., Rodrigues, R., Pereira, I.A., and Rodrigues-Pousada, C. (2013) Roles of HynAB and Ech, the only two hydrogenases found in the model sulfate reducer Desulfovibrio gigas. J. Bacteriol. 195: 4753-4760.
- Thauer, R.K., 2012. The Wolfe cycle comes full circle. Proc. Acad. Natl. Sci. USA 109: 15084-15085.
- Thauer, R.K., Kaster, A.K., Seedorf, H., Buckel, W., and Hedderich, R. (2008) Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6: 579-591.
- Welte, C., Krätzer, C., and Deppenmeier, U. (2010) Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 277:
Biochemistry:
- Forzi, L., Koch, J., Guss, A.M., Radosevich, C.G., Metcalf, W.W., and Hedderich, R. (2005) Assignment of the [4Fe-4S] clusters of Ech hydrogenase from Methanosarcina barkeri to individual subunits via the characterization of site-directed mutants. FEBS J. 272: 4741-4753.
- Kurkin, S., Meuer, J., Koch, J., Hedderich, R., and Albracht, S.P. (2002) The membrane-bound [NiFe]-hydrogenase (Ech) from Methanosarcina barkeri: unusual properties of the iron-sulphur clusters. Eur. J. Biochem. 269: 6101-6111.
- Meuer, J., Bartoschek, S., Koch, J., Künkel, A. and Hedderich, R. (1999) Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265: 325-335.
- Soboh, B., Linder, D., and Hedderich, R. (2004) A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiol. 150: 2451-2463. 3396-3403.