[NiFe] Group 4c-hydrogenase
This entry was last updated at: Feb. 9, 2018, 7:09 a.m.
Properties
| Group | [NiFe] Group 4: Respiratory H2-evolving [NiFe] hydrogenases |
| Subgroup | [NiFe] Group 4c: Carbon monoxide-respiring |
| Function | Forms respiratory supercomplex that couples CO oxidation at a carbon monoxide dehydrogenase to proton reduction at a CO-tolerant hydrogenase. The energy released is probably used to translocated protons through an unresolved mechanism. |
| Activity | H2-evolution |
| Oxygen tolerance | O2-tolerant |
| Localisation | Membrane-bound |
Distribution
| Ecosystem distribution | In low abundance in anoxic soils |
| Taxonomic distribution | Exclusive to carboxydotrophic Proteobacteria |
Genetic Organisation
e.g. Carboxydothermus hydrogenoformans (WP_011344721.1):

Architecture
| Structures |
|
| Subunits | 7 |
| Subunit description | CooH (hydrogenase large subunit) CooL (hydrogenase small subunit) CooF (polyferredoxin subunit) CooK (NuoH-like transmembrane adaptor) CooM (NuoL-like transmembrane adaptor) CooX (NuoC-like transmembrane adaptor) CooU (conserved hypothetical protein) Associates with carbon monoxide dehydrogenase |
| Catalytic site | [NiFe]-centre |
| FeS clusters | Proximal: 4Cys[4Fe4S] Medial: Absent Distal: Absent |
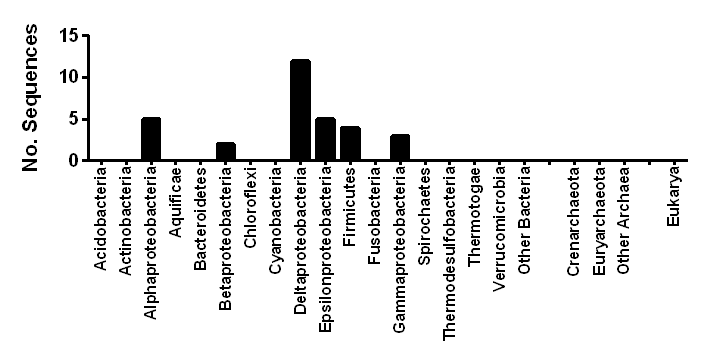
Sequences in this class
| NCBI Accession | Organism | Hydrogenase Class | Phylum | Order | Activity (Predicted) | Oxygen Tolerance (Predicted) | Subunits (Predicted) | Metal Centres (Predicted) | Accessory Subunits (Predicted) |
|---|---|---|---|---|---|---|---|---|---|
| WP_026783288.1 | Pleomorphomonas koreensis | [NiFe] Group 4c | Alphaproteobacteria | Rhizobiales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_026790609.1 | Pleomorphomonas oryzae | [NiFe] Group 4c | Alphaproteobacteria | Rhizobiales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_011474902.1 | Rhodopseudomonas palustris | [NiFe] Group 4c | Alphaproteobacteria | Rhizobiales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_008388614.1 | Rhodovulum sp. PH10 | [NiFe] Group 4c | Alphaproteobacteria | Rhodobacterales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_011389179.1 | Rhodospirillum rubrum | [NiFe] Group 4c | Alphaproteobacteria | Rhodospirillales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_028535106.1 | Paludibacterium yongneupense | [NiFe] Group 4c | Betaproteobacteria | Neisseriales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_018606884.1 | Uliginosibacterium gangwonense | [NiFe] Group 4c | Betaproteobacteria | Rhodocyclales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_009368261.1 | Bilophila sp. 4_1_30 | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_005028008.1 | Bilophila wadsworthia | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_027187553.1 | Desulfovibrio cuneatus | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_012625505.1 | Desulfovibrio desulfuricans | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_015416298.1 | Desulfovibrio piezophilus | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_006007878.1 | Desulfovibrio piger | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_027191193.1 | Desulfovibrio putealis | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_007527819.1 | Desulfovibrio sp. A2 | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_020881764.1 | Desulfovibrio sp. X2 | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_010939566.1 | Desulfovibrio vulgaris | [NiFe] Group 4c | Deltaproteobacteria | Desulfovibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_012528510.1 | Geobacter bemidjiensis | [NiFe] Group 4c | Deltaproteobacteria | Desulfuromonadales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_026842872.1 | Geobacter bremensis | [NiFe] Group 4c | Deltaproteobacteria | Desulfuromonadales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_007473061.1 | Caminibacter mediatlanticus | [NiFe] Group 4c | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_014769235.1 | Sulfurospirillum barnesii | [NiFe] Group 4c | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_012856977.1 | Sulfurospirillum deleyianum | [NiFe] Group 4c | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_025344446.1 | Sulfurospirillum multivorans | [NiFe] Group 4c | Epsilonproteobacteria | Campylobacterales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_015902767.1 | Nautilia profundicola | [NiFe] Group 4c | Epsilonproteobacteria | Nautiliales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_015594087.1 | Bacillus sp. 1NLA3E | [NiFe] Group 4c | Firmicutes | Bacillales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_013809561.1 | Desulfotomaculum carboxydivorans | [NiFe] Group 4c | Firmicutes | Clostridiales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_013121779.1 | Thermincola potens | [NiFe] Group 4c | Firmicutes | Clostridiales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_011344721.1 | Carboxydothermus hydrogenoformans | [NiFe] Group 4c | Firmicutes | ThermoanaerobacteralesCoo | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_028108808.1 | Ferrimonas futtsuensis | [NiFe] Group 4c | Gammaproteobacteria | Alteromonadales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_028112273.1 | Ferrimonas kyonanensis | [NiFe] Group 4c | Gammaproteobacteria | Alteromonadales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
| WP_007468176.1 | Photobacterium sp. AK15 | [NiFe] Group 4c | Gammaproteobacteria | Vibrionales | Evolving | Sensitive | 7 | [NiFe]-centre, 1 x [4Fe4S] clusters | NuoL, NuoH, NuoC, [FeS] protein, polyferredoxin |
Literature
Genetics:
- Bonam, D., Lehman, L., Roberts, G.P. and Ludden, P.W. (1989) Regulation of carbon monoxide dehydrogenase and hydrogenase in Rhodospirillum rubrum: effects of CO and oxygen on synthesis and activity. J. Bacteriol. 171: 3102-3107.
- Fox, J.D., He, Y., Shelver, D., Roberts, G.P., and Ludden, P.W. (1996) Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum. J. Bacteriol. 178: 6200-6208.
- Greening, C., Biswas, A., Carere, C.R., Jackson, C.J., Taylor, M.C., Stott, M.B., Cook, G.M., and Morales, S.E. (2016) Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10: 761-777.
- Kruse, S., Goris, T., Wolf, M., Wei, X., and Diekert, G. (2017). The NiFe hydrogenases of the tetrachloroethene-respiring Epsilonproteobacterium Sulfurospirillum multivorans: biochemical studies and transcription analysis. Front. Microbiol. 8: 444.
- Vanzin, G., Yu, J., Smolinski, S., Tek, V., Pennington, G., and Maness, P.C. (2010) Characterization of genes responsible for the CO-linked hydrogen production pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 76: 3715-3722.
Physiology:
- Gerhardt, M., Svetlichny, V., Sokolova, T.G., Zavarzin, G.A., and Ringpfeil, M. (1991) Bacterial CO utilization with H2 production by the strictly anaerobic lithoautotrophic thermophilic bacterium Carboxydothermus hydrogenus DSM 6008 isolated from a hot swamp. FEMS Microbiol. Lett. 83: 267-271.
- Kerby, R.L., Ludden, P.W., and Roberts, G.P. (1995) Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 177: 2241-2244.
- Maness, P.C., Huang, J., Smolinski, S., Tek, V., and Vanzin, G. (2005) Energy generation from the CO oxidation-hydrogen production pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 71: 2870-2874.
- Maness, P.C. and Weaver, P.F. (2002) Hydrogen production from a carbon-monoxide oxidation pathway in Rubrivivax gelatinosus. Int. J. Hydrogen Energy 27: 1407-1411.
- Maness, P.C., Smolinski, S., Dillon, A.C., Heben, M.J., and Weaver, P.F. (2002) Characterization of the oxygen tolerance of a hydrogenase linked to a carbon monoxide oxidation pathway in Rubrivivax gelatinosus. Appl. Environ. Microbiol. 68: 2633-2636.
- Zhao, Y., Cimpoia, R., Liu, Z., and Guiot, S.R. (2011) Kinetics of CO conversion into H2 by Carboxydothermus hydrogenoformans. Appl. Microbiol. Biotechnol. 91: 1677-1684.
Biochemistry:
- Soboh, B., Linder, D., and Hedderich, R. (2002) Purification and catalytic properties of a CO-oxidizing: H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur. J. Biochem. 269: 5712-5721.
- Walker, C.B. et al. (2009) The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J. Bacteriol. 191: 5793-5801.
- Singer, S.W., Hirst, M.B., and Ludden, P.W. (2006) CO-dependent H2 evolution by Rhodospirillum rubrum: role of CODH: CooF complex. BBA Bioenerg. 1757: 1582-1591.