[FeFe] Group A1-hydrogenase
This entry was last updated at: Feb. 9, 2018, 7:29 a.m.
Properties
| Group | [FeFe]-hydrogenases |
| Subgroup | [FeFe] Group A1: Prototypical |
| Function | Fermentative or photobiological evolution of H2 using reduced ferredoxin as an electron donor. |
| Activity | H2-evolution |
| Oxygen tolerance | O2-labile |
| Localisation | Cytosolic |
Distribution
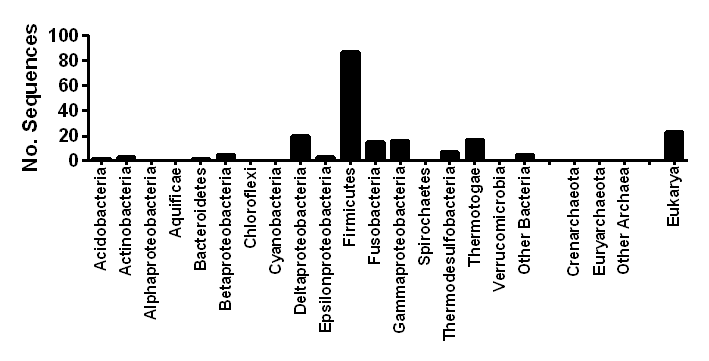
| Ecosystem distribution | Anoxic waters, soils, and sediments, gastrointestinal tracts |
| Taxonomic distribution | Widespread in anaerobic bacteria and unicellular eukaryotes |
Genetic Organisation
Organisation 1: Chlamydomonas reinhardtii (XP_001693376.1):

Organisation 2: Trichomonas vaginalis (XP_001330775.1):

Organisation 3: Clostridium pasteurianum (WP_004455619.1):

Organisation 4: Desulfovibrio vulgaris (WP_010939057.1):

Organisation 5: Shewanella oneidensis (WP_011073665.1):
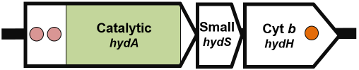
Organisation 6: Trichomonas vaginalis (XP_001580488.1):

Architecture
| Structures | |
| Subunits | N/A |
| Subunit description | HydA (catalytic subunit) In organisation 4 and 5, the catalytic subunit is split into a large subunit (HydA) and small subunit (HydS) In organisation 5, a cytochrome b subunit (HydH) is present In organisation 6, the hydrogenase domain is fused to a nitrate reductase or sulfite reductase domain |
| Catalytic site | [FeFe]-centre |
| FeS clusters | Organisation 1: Absent Organisation 2: 2 [4Fe4S] clusters Organisation 3: 3 [4Fe4S] clusters, 1 [2Fe2S] cluster Organisation 4: 2 [4Fe4S] clusters Organisation 5: 2 [4Fe4S] clusters Organisation 6: 3 [4Fe4S] clusters, 1 [2Fe2S] cluster |
Sequences in this class
| NCBI Accession | Organism | Hydrogenase Class | Phylum | Order | Activity (Predicted) | Oxygen Tolerance (Predicted) | Subunits (Predicted) | Metal Centres (Predicted) | Accessory Subunits (Predicted) |
|---|---|---|---|---|---|---|---|---|---|
| XP_001322682.1 | Trichomonas vaginalis | [FeFe] Group A1 | Metamonada | Trichomonadida | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | Fusion with ferredoxin reductase |
| XP_001580488.1 | Trichomonas vaginalis | [FeFe] Group A1 | Metamonada | Trichomonadida | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | Fusion with sulfite reductase |
| WP_002568840.1 | Lachnoclostridium bolteae | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_002583070.1 | Clostridium butyricum | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_002597295.1 | Clostridium colicanis | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_002599108.1 | Clostridium colicanis | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_002599284.1 | Clostridium colicanis | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_002607524.1 | Erysipelatoclostridium innocuum | [FeFe] Group A1 | Firmicutes | Erysipelotrichales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_002945014.1 | Campylobacter rectus | [FeFe] Group A1 | Epsilonproteobacteria | Campylobacterales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_003514900.1 | Ruminiclostridium thermocellum | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_004455619.1 | Clostridium pasteurianum | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_004607406.1 | Lachnoclostridium scindens | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_004697562.1 | Veillonella parvula | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005026529.1 | Bilophila wadsworthia | [FeFe] Group A1 | Deltaproteobacteria | Desulfovibrionales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005035184.1 | Holophaga foetida | [FeFe] Group A1 | Acidobacteria | Holophagales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_005038203.1 | Holophaga foetida | [FeFe] Group A1 | Acidobacteria | Holophagales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005351469.1 | Eubacterium hallii | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005375825.1 | Veillonella atypica | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005378576.1 | Veillonella atypica | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005382858.1 | Veillonella atypica | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005384713.1 | Veillonella atypica | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005384882.1 | Veillonella dispar | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005385706.1 | Veillonella dispar | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005436344.1 | Sutterella wadsworthensis | [FeFe] Group A1 | Betaproteobacteria | Burkholderiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005884626.1 | Fusobacterium mortiferum | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005886368.1 | Fusobacterium mortiferum | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005952086.1 | Fusobacterium varium | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005957191.1 | Fusobacterium necrophorum | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_005958327.1 | Fusobacterium necrophorum | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005976079.1 | Fusobacterium ulcerans | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_005981915.1 | Fusobacterium ulcerans | [FeFe] Group A1 | Fusobacteria | Fusobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006362914.1 | Slackia exigua | [FeFe] Group A1 | Actinobacteria | Coriobacteriales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006442921.1 | Lachnoclostridium hylemonae | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006492050.1 | Mesotoga infera | [FeFe] Group A1 | Thermotogae | Thermotogales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_006556240.1 | Veillonella ratti | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006566021.1 | Anaerostipes caccae | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006571360.1 | Pseudoflavonifractor capillosus | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006627482.1 | Bulleidia extructa | [FeFe] Group A1 | Firmicutes | Erysipelotrichales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006780212.1 | Lachnoclostridium hathewayi | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006789933.1 | Anaeroglobus geminatus | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_006942403.1 | Megasphaera micronuciformis | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_007285570.1 | Intestinibacter bartlettii | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_007716914.1 | Lachnoclostridium asparagiforme | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_007866252.1 | Lachnoclostridium citroniae | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_007951362.1 | Pelosinus fermentans | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_008193913.1 | Thermotoga sp. EMP | [FeFe] Group A1 | Thermotogae | Thermotogales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_008412697.1 | Desulfotomaculum hydrothermale | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_009145476.1 | Phascolarctobacterium succinatutens | [FeFe] Group A1 | Firmicutes | Selenomonadales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
| WP_010074109.1 | Clostridium cellulovorans | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 3 x [4Fe4S] clusters, 1 x [2Fe2S] cluster | None |
| WP_010234442.1 | Clostridium arbusti | [FeFe] Group A1 | Firmicutes | Clostridiales | Evolving | Labile | 1 | [FeFe]-centre, 2 x [4Fe4S] clusters | None |
Literature
Genetics:
- Atta, M. and Meyer, J. (2000) Characterization of the gene encoding the [Fe]-hydrogenase from Megasphaera elsdenii. BBA Prot. Struct. Mol. Enzymol. 1476: 368-371.
- Baba, R., Morita, M., Asakawa, S., and Watanabe, T. (2017) Transcription of [FeFe]-hydrogenase genes during H2 production in Clostridium and Desulfovibrio spp. isolated from a paddy field soil. Microb. Environ. 32: 125-132.
- Calusinska, M., Happe, T., Joris, B., and Wilmotte, A. (2010) The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiol. 156: 1575-1588.
- Forestier, M., King, P., Zhang, L., Posewitz, M., Schwarzer, S., Happe, T., Ghirardi, M.L., and Seibert, M. (2003) Expression of two [Fe]-hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Eur. J. Biochem. 270: 2750-2758.
- Gorwa, M.F., Croux, C., and Soucaille, P. (1996) Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824. J. Bacteriol. 178: 2668-2675.
- Greening, C., Biswas, A., Carere, C.R., Jackson, C.J., Taylor, M.C., Stott, M.B., Cook, G.M., and Morales, S.E. (2016) Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 10: 761-777.
- Happe, T. and Kaminski, A. (2002) Differential regulation of the Fe-hydrogenase during anaerobic adaptation in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 269: 1022-1032.
- Happe, T. and Naber, J.D. (1993) Isolation, characterization and N-terminal amino acid sequence of hydrogenase from the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214: 475-481.
- Horner, D.S., Foster, P.G. and Embley, T.M. (2000) Iron hydrogenases and the evolution of anaerobic eukaryotes. Mol. Biol. Evol. 17: 1695-1709.
- Kuchenreuther, J.M., Grady-Smith, C.S., Bingham, A.S., George, S.J., Cramer, S.P., and Swartz, J.R. (2010) High-yield expression of heterologous [FeFe] hydrogenases in Escherichia coli. PLOS One 5: e15491.
- Meyer, J. and Gagnon, J. (1991) Primary structure of hydrogenase from Clostridium pasteurianum. Biochem. 30: 9697-9704.
- Poudel, S., Tokmina-Lukaszewska, M., Colman, D.R., Refai, M., Schut, G.J., King, P.W., Maness, P.C., Adams, M.W., Peters, J.W., Bothner, B., and Boyd, E.S. (2016) Unification of [FeFe]-hydrogenases into three structural and functional groups. BBA Gen. Subj. 1890: 1910-1921.
- Stapleton, J.A. and Swartz, J.R. (2010) Development of an in vitro compartmentalization screen for high-throughput directed evolution of [FeFe] hydrogenases. PLOS One 5: e15275.
- Therien, J.B., Artz, J.H., Poudel, S., Hamilton, T.L., Liu, Z., Noone, S.M., Adams, M.W., King, P.W., Bryant, D.A., Boyd, E.S., and Peters, J.W. (2017) The physiological functions and structural determinants of catalytic bias in the [FeFe]-hydrogenases CpI and CpII of Clostridium pasteurianum strain W5. Front. Microbiol. 8: 1305.
- Voncken, F.G., Boxma, B., van Hoek, A.H., Akhmanova, A.S., Vogels, G.D., Huynen, M., Veenhuis, M., and Hackstein, J.H. (2002) A hydrogenosomal [Fe]-hydrogenase from the anaerobic chytrid Neocallimastix sp. L2. Gene 284: 103-112.
Physiology:
- Cornish, A.J., Gärtner, K., Yang, H., Peters, J.W., and Hegg, E.L. (2011) Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum. J. Biol. Chem. 286: 38341-38347.
- Drake, H.L. (1982) Demonstration of hydrogenase in extracts of the homoacetate-fermenting bacterium Clostridium thermoaceticum. J. Bacteriol. 150: 702-709.
- Florin, L., Tsokoglou, A., and Happe, T. (2001) A novel type of iron hydrogenase in the green alga Scenedesmus obliquus is linked to the photosynthetic electron transport chain. J. Biol. Chem. 276: 6125-6132.
- Fouchard, S., Hemschemeier, A., Caruana, A., Pruvost, J., Legrand, J., Happe, T., Peltier, G., and Cournac, L. (2005) Autotrophic and mixotrophic hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. Appl. Environ. Microbiol. 71: 6199-6205.
- Hamann, E. et al. (2016) Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature 534: 254-258.
- Happe, T., Mosler, B., and Naber, J.D. (1994) Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222: 769-774.
- Hemschemeier, A., Fouchard, S., Cournac, L., Peltier, G., and Happe, T. (2008) Hydrogen production by Chlamydomonas reinhardtii: an elaborate interplay of electron sources and sinks. Planta 227: 397-407.
- Kelly, C.L., Pinske, C., Murphy, B.J., Parkin, A., Armstrong, F., Palmer, T., and Sargent, F. (2015) Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli. Biotech. Rep. 8: 94-104.
- Koo, J., Shigi, S., Rohovie, M., Mehta, K., and Swartz, J.R. (2016) Characterization of [FeFe] hydrogenase O2 sensitivity using a new, physiological approach. J. Biol. Chem. 291: 21563-21570.
- Lemon, B.J. and Peters, J.W. (1999) Binding of exogenously added carbon monoxide at the active site of the iron-only hydrogenase (CpI) from Clostridium pasteurianum. Biochem. 38: 12969-12973.
- Meuser, J.E., D'Adamo, S., Jinkerson, R.E., Mus, F., Yang, W., Ghirardi, M.L., Seibert, M., Grossman, A.R., and Posewitz, M.C. (2012) Genetic disruption of both Chlamydomonas reinhardtii [FeFe]-hydrogenases: insight into the role of HYDA2 in H2 production. Biochem. Biophys. Res. Commun. 417: 704-709.
- Nicolet, Y., Lemon, B.J., Fontecilla-Camps, J.C., and Peters, J.W. (2000) A novel FeS cluster in Fe-only hydrogenases. Trends Biochem. Sci. 25: 138-143.
- Noth, J., Krawietz, D., Hemschemeier, A., and Happe, T. (2013) Pyruvate: ferredoxin oxidoreductase is coupled to light-independent hydrogen production in Chlamydomonas reinhardtii. J. Biol. Chem. 288: 4368-4377.
- Peck Jr, H.D. and Gest, H. (1957) Hydrogenase of Clostridium butylicum. J. Bacteriol. 73: 569-578.
- Sawyer, A., Bai, Y., Lu, Y., Hemschemeier, A., and Happe, T. (2017) Compartmentalisation of [FeFe]‐hydrogenase maturation in Chlamydomonas reinhardtii. Plant J. 90: 1134-1143.
- Shaw, A.J., Hogsett, D.A., and Lynd, L.R. (2009) Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J. Bacteriol. 191: 6457-6464.
- Tolleter, D. et al. (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23: 2619-2630.
- Winkler, M., Heil, B., Heil, B. and Happe, T. (2002) Isolation and molecular characterization of the [Fe]-hydrogenase from the unicellular green alga Chlorella fusca. BBA Gene Struct. Express. 1576: 330-334.
- Yacoby, I., Pochekailov, S., Toporik, H., Ghirardi, M.L., King, P.W., and Zhang, S. (2011) Photosynthetic electron partitioning between [FeFe]-hydrogenase and ferredoxin: NADP+-oxidoreductase (FNR) enzymes in vitro. Proc. Natl. Acad. Sci. USA 108: 9396-9401.
- Zhang, L., Happe, T., and Melis, A. (2002) Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214: 552-561.
Biochemistry:
- Adams, M.W. (1987) The mechanisms of H2 activation and CO binding by hydrogenase I and hydrogenase II of Clostridium pasteurianum. J. Biol. Chem. 262: 15054-15061.
- Adams, M.W., Eccleston, E., and Howard, J.B. (1989) Iron-sulfur clusters of hydrogenase I and hydrogenase II of Clostridium pasteurianum. Proc. Natl. Acad. Sci. USA 86: 4932-4936.
- Adams, M.W. and Mortenson, L.E. (1984) The purification of hydrogenase II (uptake hydrogenase) from the anaerobic N2-fixing bacterium Clostridium pasteurianum. BBA Bioenerg. 766: 51-61.
- Adams, M.W. and Mortenson, L.E. (1984) The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. A comparison with hydrogenase I. J. Biol. Chem. 259: 7045-7055.
- Artz, J.H., Mulder, D.W., Ratzloff, M.W., Lubner, C.E., Zadvornyy, O.A., LeVan, A.X., Williams, S.G., Adams, M.W., Jones, A.K., King, P.W., and Peters, J.W. (2017) Reduction potentials of [FeFe]-hydrogenase accessory iron–sulfur clusters provide insights into the energetics of proton reduction catalysis. J. Am. Chem. Soc. 139: 9544-9550.
- Bennett, B., Lemon, B.J., and Peters, J.W. (2000) Reversible carbon monoxide binding and inhibition at the active site of the Fe-only hydrogenase. Biochem. 39: 7455-7460.
- Berlier, Y., Fauque, G.D., LeGall, J., Choi, E.S., Peck, H.D., and Lespinat, P.A. (1987) Inhibition studies of three classes of Desulfovibrio hydrogenase: application to the further characterization of the multiple hydrogenases found in Desulfovibrio vulgaris Hildenborough. Biochem. Biophys. Res. Commun. 146: 147-153.
- Bingham, A.S., Smith, P.R., and Swartz, J.R. (2012) Evolution of an [FeFe] hydrogenase with decreased oxygen sensitivity. Int. J. Hydrogen Energy 37: 2965-2976.
- Brown, K.A., Wilker, M.B., Boehm, M., Dukovic, G., and King, P.W. (2012) Characterization of photochemical processes for H2 production by CdS nanorod [FeFe] hydrogenase complexes. J. Am. Chem. Soc. 134: 5627-5636.
- Chen, J.S. and Blanchard, D.K. (1978) Isolation and properties of a unidirectional H2-oxidizing hydrogenase from the strictly anaerobic N2-fixing bacterium Clostridium pasteurianum W5. Biochem. Biophys. Res. Commun. 84: 1144-1150.
- Chen, J.S. and Mortenson, L.E. (1974) Purification and properties of hydrogenase from Clostridium pasteurianum W5. BBA Prot. Struct. 371: 283-298.
- Cohen, J., Kim, K., King, P., Seibert, M., and Schulten, K. (2005) Finding gas diffusion pathways in proteins: application to O2 and H2 transport in CpI [FeFe]-hydrogenase and the role of packing defects. Structure 13: 1321-1329.
- Demuez, M., Cournac, L., Guerrini, O., Soucaille, P., and Girbal, L. (2007) Complete activity profile of Clostridium acetobutylicum [FeFe]-hydrogenase and kinetic parameters for endogenous redox partners. FEMS Microbiol. Lett. 275: 113-121.
- Dijk, C., Grande, H.J., Mayhew, S.G., and Veeger, C (1980) Properties of the hydrogenase of Megasphaera elsdenii. Eur. J. Biochem. 107: 251-261.
- Dijk, C., Mayhew, S.G., Grande, H.J., and Veeger, C. (1979) Purification and properties of hydrogenase from Megasphaera elsdenii. Eur. J. Biochem. 102: 317-330.
- Dijk, C. and Veeger, C. (1981) The effects of pH and redox potential on the hydrogen production activity of the hydrogenase from Megasphaera elsdenii. Eur. J. Biochem. 114: 209-219.
- Engelbrecht, V., Rodríguez-Maciá, P., Esselborn, J., Sawyer, A., Hemschemeier, A., Rüdiger, O., Lubitz, W., Winkler, M., and Happe, T. (2017) The structurally unique photosynthetic Chlorella variabilis NC64A hydrogenase does not interact with plant-type ferredoxins. BBA Bioenerg. 1858: 771-778.
- Erbes, D.L., Burris, R.H., and Orme-Johnson, W.H. (1975) On the iron-sulfur cluster in hydrogenase from Clostridium pasteurianum W5. Proc. Natl. Acad. Sci. USA 72: 4795-4799.
- Erbes, D.L. and Burris, R.H. (1978) The kinetics of methyl viologen oxidation and reduction by the hydrogenase from Clostridium pasteurianium. BBA Enzymol. 525: 45-54.
- Filipiak, M., Hagen, W.R., and Veeger, C. (1989) Hydrodynamic, structural and magnetic properties of Megasphaera elsdenii Fe hydrogenase reinvestigated. Eur. J. Biochem. 185: 547-553.
- Fourmond, V. et al. (2014) The oxidative inactivation of FeFe hydrogenase reveals the flexibility of the H-cluster. Nat. Chem. 6: 336-342.
- Gauquelin, C., Baffert, C., Richaud, P., Kamionka, E., Etienne, E., Guieysse, D., Girbal, L., Fourmond, V., André, I., Guigliarelli, B., and Léger, C. (2018) Roles of the F-domain in [FeFe] hydrogenase. BBA Bioenerg. 1859: 69-77.
- Girbal, L., von Abendroth, G., Winkler, M., Benton, P.M., Meynial-Salles, I., Croux, C., Peters, J.W., Happe, T., and Soucaille, P. (2005) Homologous and heterologous overexpression in Clostridium acetobutylicum and characterization of purified clostridial and algal Fe-only hydrogenases with high specific activities. Appl. Environ. Microbiol. 71: 2777-2781.
- Knörzer, P., Silakov, A., Foster, C.E., Armstrong, F.A., Lubitz, W., and Happe, T. (2012) Importance of the protein framework for catalytic activity of [FeFe]-hydrogenases. J. Biol. Chem. 287: 1489-1499.
- Kubas, A., Orain, C., De Sancho, D., Saujet, L., Sensi, M., Gauquelin, C., Meynial-Salles, I., Soucaille, P., Bottin, H., Baffert, C., and Fourmond, V. (2017) Mechanism of O2 diffusion and reduction in FeFe hydrogenases. Nature Chem. 9: 88-95.
- Lambertz, C., Leidel, N., Havelius, K.G., Noth, J., Chernev, P., Winkler, M., Happe, T., and Haumann, M. (2011) O2 reactions at the six-iron active site (H-cluster) in [FeFe]-hydrogenase. J. Biol. Chem. 286: 40614-40623.
- Lemon, B.J. and Peters, J.W. (1999) Binding of exogenously added carbon monoxide at the active site of the iron-only hydrogenase (CpI) from Clostridium pasteurianum. Biochem. 38: 12969-12973.
- Lubner, C.E., Knörzer, P., Silva, P.J., Vincent, K.A., Happe, T., Bryant, D.A., and Golbeck, J.H. (2010) Wiring an [FeFe]-hydrogenase with photosystem I for light-induced hydrogen production. Biochem. 49: 10264-10266.
- Morra, S., Arizzi, M., Valetti, F., and Gilardi, G. (2016) Oxygen stability in the new [FeFe]-hydrogenase from Clostridium beijerinckii SM10 (CbA5H). Biochem. 55: 5897-5900.
- Mulder, D.W., Shepard, E.M., Meuser, J.E., Joshi, N., King, P.W., Posewitz, M.C., Broderick, J.B., and Peters, J.W. (2011) Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure 19: 1038-1052.
- Hambourger, M., Gervaldo, M., Svedruzic, D., King, P.W., Gust, D., Ghirardi, M., Moore, A.L., and Moore, T.A. (2008) [FeFe]-hydrogenase-catalyzed H2 production in a photoelectrochemical biofuel cell. J. Am. Chem. Soc. 130: 2015-2022.
- Nicolet, Y., Piras, C., Legrand, P., Hatchikian, C.E., and Fontecilla-Camps, J.C. (1999) Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure 7: 13-23.
- Nicolet, Y., de Lacey, A.L., Vernede, X., Fernandez, V.M., Hatchikian, E.C., and Fontecilla-Camps, J.C., 2001. Crystallographic and FTIR spectroscopic evidence of changes in Fe coordination upon reduction of the active Site of the Fe-only Hydrogenase from Desulfovibrio desulfuricans. J. Am. Chem. Soc. 123: 1596-1601.
- Payne, M.J., Chapman, A., and Cammack, R. (1993) Evidence for an [Fe]-type hydrogenase in the parasitic protozoan Trichomonas vaginalis. FEBS Lett. 317: 101-104.
- Peters, J.W., Lanzilotta, W.N., Lemon, B.J., and Seefeldt, L.C. (1998) X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282: 1853-1858.
- Philipps, G., Happe, T., and Hemschemeier, A. (2012) Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 235: 729-745.
- Plumeré, N., Rüdiger, O., Oughli, A.A., Williams, R., Vivekananthan, J., Pöller, S., Schuhmann, W., and Lubitz, W. (2014) A redox hydrogel protects hydrogenase from high-potential deactivation and oxygen damage. Nat. Chem. 6: 822-827.
- Sawyer, A. and Winkler, M. (2017) Evolution of Chlamydomonas reinhardtii ferredoxins and their interactions with [FeFe]-hydrogenases. Photosynth. Res. 134: 307-316.
- Stripp, S.T., Goldet, G., Brandmayr, C., Sanganas, O., Vincent, K.A., Haumann, M., Armstrong, F.A., and Happe, T. (2009) How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl. Acad. Sci. USA 106: 17331-17336.
- Winkler, M., Kuhlgert, S., Hippler, M., and Happe, T. (2009) Characterization of the key step for light-driven hydrogen evolution in green algae. J. Biol. Chem. 284: 36620-36627.
Chemistry:
- Adamska, A., Silakov, A., Lambertz, C., Rödiger, O., Happe, T., Reijerse, E., and Lubitz, W. (2012) Identification and characterization of the "super-reduced" state of the H-Cluster in [FeFe] hydrogenase: a new building block for the catalytic cycle? Angewandte Chemie 51: 11458-11462.
- Adamska-Venkatesh, A., Krawietz, D., Siebel, J., Weber, K., Happe, T., Reijerse, E., and Lubitz, W. (2014) New redox states observed in [FeFe] hydrogenases reveal redox coupling within the H-cluster. J. Am. Chem. Soc. 136: 11339-11346.
- Albracht, S.P., Roseboom, W., and Hatchikian, E.C. (2006) The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. I. Light sensitivity and magnetic hyperfine interactions as observed by electron paramagnetic resonance. J. Biol. Inorg. Chem. 11: 88-101.
- Berggren, G., Adamska, A., Lambertz, C., Simmons, T.R., Esselborn, J., Atta, M., Gambarelli, S., Mouesca, J.M., Reijerse, E., Lubitz, W., and Happe, T. (2013) Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature 499: 66-69.
- Bruschi, M., Greco, C., Bertini, L., Fantucci, P., Ryde, U., and Gioia, L.D. (2010) Functionally relevant interplay between the Fe4S4 cluster and CN- ligands in the active site of [FeFe]-hydrogenases. J. Am. Chem. Soc. 132: 4992-4993.
- Bruschi, M., Greco, C., Kaukonen, M., Fantucci, P., Ryde, U., and De Gioia, L. (2009) Influence of the [2Fe] H subcluster environment on the properties of key intermediates in the catalytic cycle of [FeFe] hydrogenases: hints for the rational design of synthetic catalysts. Angewandte Chemie 48: 3503-3506.
- Butt, J.N., Filipiak, M., and Hagen, W.R. (1997) Direct electrochemistry of Megasphaera elsdenii iron hydrogenase. Eur. J. Biochem. 245: 116-122.
- Chen, Z., Lemon, B.J., Huang, S., Swartz, D.J., Peters, J.W., and Bagley, K.A. (2002) Infrared studies of the CO-inhibited form of the Fe-only hydrogenase from Clostridium pasteurianum I: examination of its light sensitivity at cryogenic temperatures. Biochem. 41: 2036-2043.
- Cornish, A.J., Gärtner, K., Yang, H., Peters, J.W., and Hegg, E.L. (2011) Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum. J. Biol. Chem. 286: 38341-38347.
- De Lacey, A.L., Stadler, C., Cavazza, C., Hatchikian, E.C., and Fernandez, V.M. (2000) FTIR characterization of the active site of the Fe-hydrogenase from Desulfovibrio desulfuricans. J. Am. Chem. Soc. 122: 11232-11233.
- Foster, C.E., Krämer, T., Wait, A.F., Parkin, A., Jennings, D.P., Happe, T., McGrady, J.E., and Armstrong, F.A. (2012) Inhibition of [FeFe]-hydrogenases by formaldehyde and wider mechanistic implications for biohydrogen activation. J. Am. Chem. Soc. 134: 7553-7557.
- Grande, H.J., Dunham, W.R., Averill, B., Dijk, C., and Sands, R.H. (1983) Electron paramagnetic resonance and other properties of hydrogenases isolated from Desulfovibrio vulgaris (strain Hildenborough) and Megasphaera elsdenii. Eur. J. Biochem. 136: 201-207.
- Greco, C., Bruschi, M., Heimdal, J., Fantucci, P., De Gioia, L., and Ryde, U. (2007) Structural insights into the active-ready form of [FeFe]-hydrogenase and mechanistic details of its inhibition by carbon monoxide. Inorg. Chem. 46: 7256-7258.
- Kamp, C., Silakov, A., Winkler, M., Reijerse, E.J., Lubitz, W., and Happe, T. (2008) Isolation and first EPR characterization of the [FeFe]-hydrogenases from green algae. BBA Bioenerg. 1777: 410-416.
- Lemon, B.J. and Peters, J.W. (2000) Photochemistry at the active site of the carbon monoxide inhibited form of the iron-only hydrogenase (CpI). J. Am. Chem. Soc. 122: 3793-3794.
- Madden, C., Vaughn, M.D., Díez-Pérez, I., Brown, K.A., King, P.W., Gust, D., Moore, A.L., and Moore, T.A. (2011) Catalytic turnover of [FeFe]-hydrogenase based on single-molecule imaging. J. Am. Chem. Soc. 134: 1577-1582.
- Mebs, S., Kositzki, R., Duan, J., Kertess, L., Senger, M., Wittkamp, F., Apfel, U.P., Happe, T., Stripp, S.T., Winkler, M., and Haumann, M. (2018) Hydrogen and oxygen trapping at the H-cluster of [FeFe]-hydrogenase revealed by site-selective spectroscopy and QM/MM calculations. BBA Bioenerg. 1859: 28-41.
- Mebs, S., Senger, M., Duan, J., Wittkamp, F., Apfel, U.P., Happe, T., Winkler, M., Stripp, S.T., and Haumann, M. (2017) Bridging hydride at reduced H-cluster species in [FeFe]-hydrogenases revealed by infrared spectroscopy, isotope editing, and quantum chemistry. J. Am. Chem. Soc. 139: 12157-12160.
- Mirmohades, M., Adamska-Venkatesh, A., Sommer, C., Reijerse, E.J., Lomoth, R., Lubitz, W., and Hammarström, L. (2016) Following [FeFe] hydrogenase active site intermediates by time-resolved mid-IR spectroscopy. J. Phys. Chem. Lett. 7: 3290-3293.
- Mulder, D.W., Guo, Y., Ratzloff, M.W., and King, P.W. (2016) Identification of a catalytic iron-hydride at the H-cluster of [FeFe]-hydrogenase. J. Am. Chem. Soc. 139: 83-86.
- Mulder, D.W., Ratzloff, M.W., Shepard, E.M., Byer, A.S., Noone, S.M., Peters, J.W., Broderick, J.B., and King, P.W. (2013) EPR and FTIR analysis of the mechanism of H2 activation by [FeFe]-hydrogenase HydA1 from Chlamydomonas reinhardtii. J. Am. Chem. Soc. 135: 6921-6929.
- Nicolet, Y., de Lacey, A.L., Vernede, X., Fernandez, V.M., Hatchikian, E.C., and Fontecilla-Camps, J.C. (2001) Crystallographic and FTIR spectroscopic evidence of changes in Fe coordination upon reduction of the active site of the Fe-only hydrogenase from Desulfovibrio desulfuricans. J. Am. Chem. Soc. 123: 1596-1601.
- Pandey, K., Islam, S.T., Happe, T., and Armstrong, F.A. (2017) Frequency and potential dependence of reversible electrocatalytic hydrogen interconversion by [FeFe]-hydrogenases. Proc. Natl. Acad. Sci. USA 114: 3843-3848.
- Parkin, A., Cavazza, C., Fontecilla-Camps, J.C., and Armstrong, F.A. (2006) Electrochemical investigations of the interconversions between catalytic and inhibited states of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. J. Am. Chem. Soc. 128: 16808-16815.
- Reijerse, E.J., Pham, C.C., Pelmenschikov, V., Gilbert-Wilson, R., Adamska-Venkatesh, A., Siebel, J.F., Gee, L.B., Yoda, Y., Tamasaku, K., Lubitz, W., and Rauchfuss, T.B. (2017) Direct observation of an iron-bound terminal hydride in [FeFe]-hydrogenase by nuclear resonance vibrational spectroscopy. J. Am. Chem. Soc. 139: 4306-4309.
- Rodríguez-Maciá, P., Pawlak, K., Rüdiger, O., Reijerse, E.J., Lubitz, W., and Birrell, J.A. (2017) Intercluster redox coupling influences protonation at the H-cluster in [FeFe] hydrogenases. J. Am. Chem. Soc. 139: 15122-15134.
- Roseboom, W., De Lacey, A.L., Fernandez, V.M., Hatchikian, E.C. and Albracht, S.P. (2006) The active site of the [FeFe]-hydrogenase from Desulfovibrio desulfuricans. II. Redox properties, light sensitivity and CO-ligand exchange as observed by infrared spectroscopy. J. Biol. Inorg. Chem. 11: 102-118.
- Ryde, U., Greco, C., and De Gioia, L. (2010) Quantum refinement of [FeFe] hydrogenase indicates a dithiomethylamine ligand. J. Am. Chem. Soc. 132: 4512-4513.
- Silakov, A., Kamp, C., Reijerse, E., Happe, T., and Lubitz, W. (2009) Spectroelectrochemical characterization of the active site of the [FeFe] hydrogenase HydA1 from Chlamydomonas reinhardtii. Biochem. 48: 7780-7786.
- Silakov, A., Reijerse, E.J., Albracht, S.P., Hatchikian, E.C., and Lubitz, W. (2007) The electronic structure of the H-cluster in the [FeFe]-hydrogenase from Desulfovibrio desulfuricans: a Q-band 57Fe-ENDOR and HYSCORE study. J. Am. Chem. Soc. 129: 11447-11458.
- Silakov, A., Wenk, B., Reijerse, E. and Lubitz, W. (2009) 14N HYSCORE investigation of the H-cluster of [FeFe] hydrogenase: evidence for a nitrogen in the dithiol bridge. Phys. Chem. Chem. Phys. 11: 6592-6599.
- Senger, M., Mebs, S., Duan, J., Shulenina, O., Laun, K., Kertess, L., Wittkamp, F., Apfel, U.P., Happe, T., Winkler, M., and Haumann, M. (2018) Protonation/reduction dynamics at the [4Fe4S] cluster of the hydrogen-forming cofactor in [FeFe]-hydrogenases. Phys. Chem. Chem. Phys. 20: 3128-3140.
- Sommer, C., Adamska-Venkatesh, A., Pawlak, K., Birrell, J.A., Rüdiger, O., Reijerse, E.J., and Lubitz, W. (2017) Proton coupled electronic rearrangement within the H-cluster as an essential step in the catalytic cycle of [FeFe] hydrogenases. J. Am. Chem. Soc. 139: 1440-1443.
- Stiebritz, M.T. and Reiher, M. (2009) Theoretical study of dioxygen induced inhibition of [FeFe]-hydrogenase. Inorg. Chem. 48: 7127-7140.
- Stripp, S., Sanganas, O., Happe, T., and Haumann, M. (2009) The structure of the active site H-cluster of [FeFe] hydrogenase from the green alga Chlamydomonas reinhardtii studied by X-ray absorption spectroscopy. Biochem. 48: 5042-5049.
- Swanson, K.D., Ratzloff, M.W., Mulder, D.W., Artz, J.H., Ghose, S., Hoffman, A., White, S., Zadvornyy, O.A., Broderick, J.B., Bothner, B., and King, P.W. (2015) [FeFe]-hydrogenase oxygen inactivation is initiated at the H cluster 2Fe subcluster. J. Am. Chem. Soc. 137: 1809-1816.
- Winkler, M., Senger, M., Duan, J., Esselborn, J., Wittkamp, F., Hofmann, E., Apfel, U.P., Stripp, S.T., and Happe, T. (2017) Accumulating the hydride state in the catalytic cycle of [FeFe]-Hydrogenases. Nature Comm. 8: 16115.